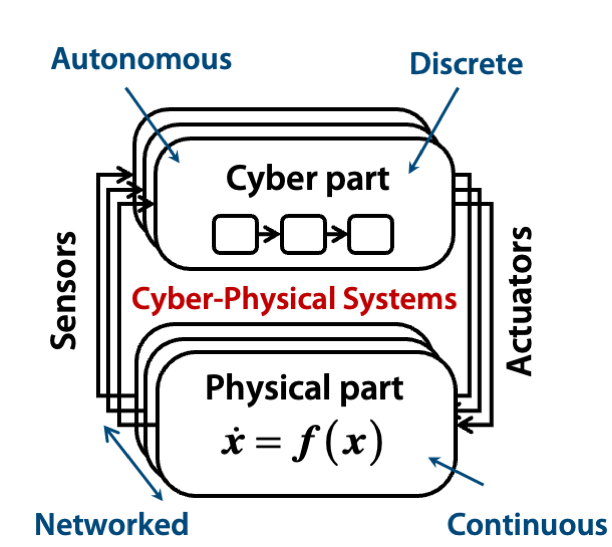

At the core of my work is the challenge of verifying and validating systems where traditional model-driven approaches may fall short due to oversimplifications and the increasing incorporation of data-driven components. My research addresses these challenges by integrating runtime verification, formal specification languages, and monitoring techniques into the system design process, ensuring more robust assurance in dynamic environments. A key area of my contribution is the analysis of probabilistic programs, where I co-developed tools such as:
|
Impressum
| Name: | Univ.-Prof. Dott. Ric. Ezio Bartocci | Title: | University Professor in Computer Science | |
| Affiliation: | Institute of Computer Engineering TU Wien – Technische Universität Wien |
Address: | TU Wien, Treitlstrasse 3, 1040 Vienna, Austria | |
| Email: | ezio.bartocci@tuwien.ac.at | Responsible for Content: | Ezio Bartocci, § 55 Abs. 2 RStV |
Disclaimer: I assume no liability for the content of external links. Responsibility lies solely with the operators of linked websites. Copyright: All content on this site is © Ezio Bartocci unless otherwise noted. Reuse without permission is not allowed. Privacy Policy (Datenschutzerklärung): This website does not collect personal data, use cookies, or analytics services.